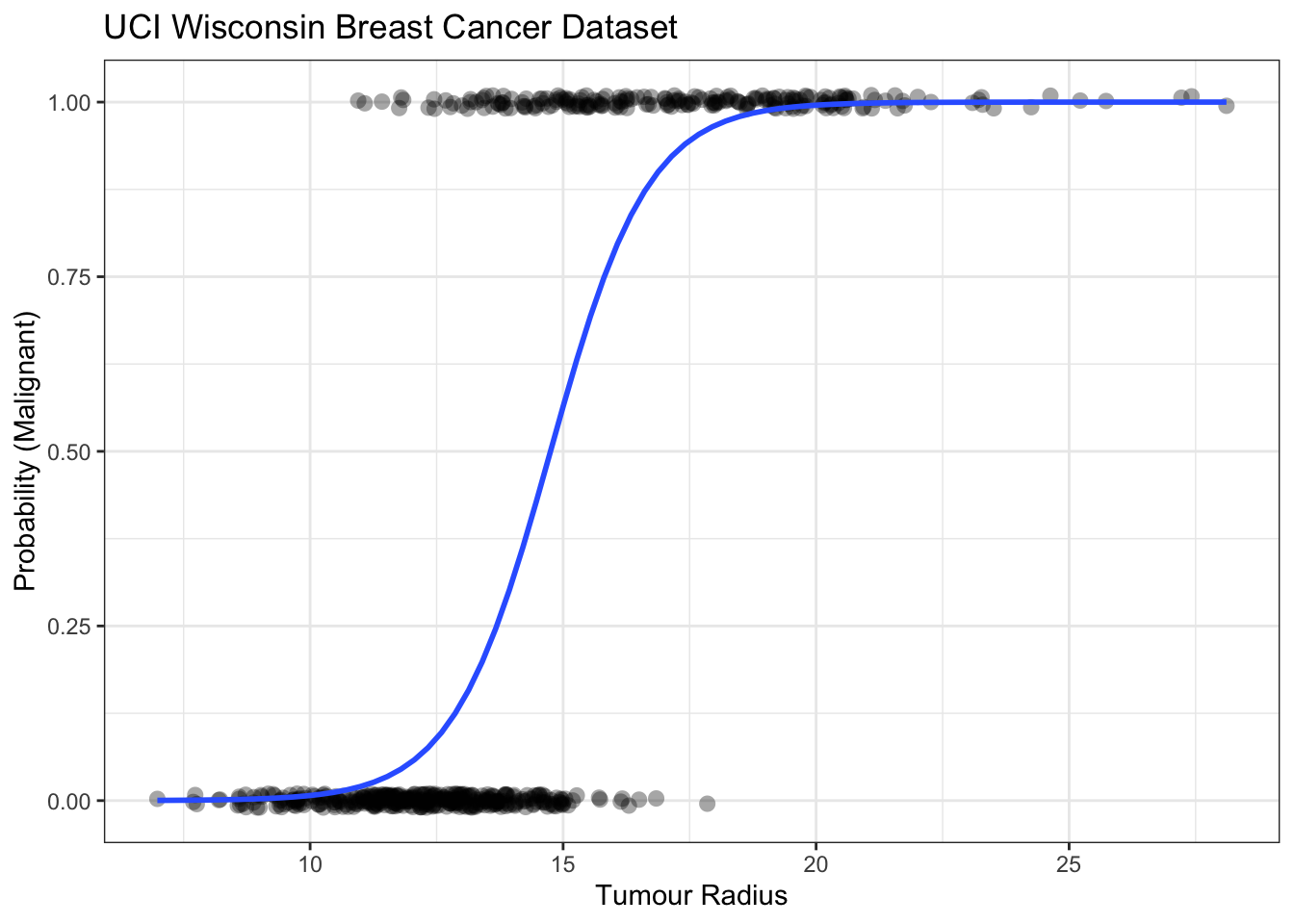
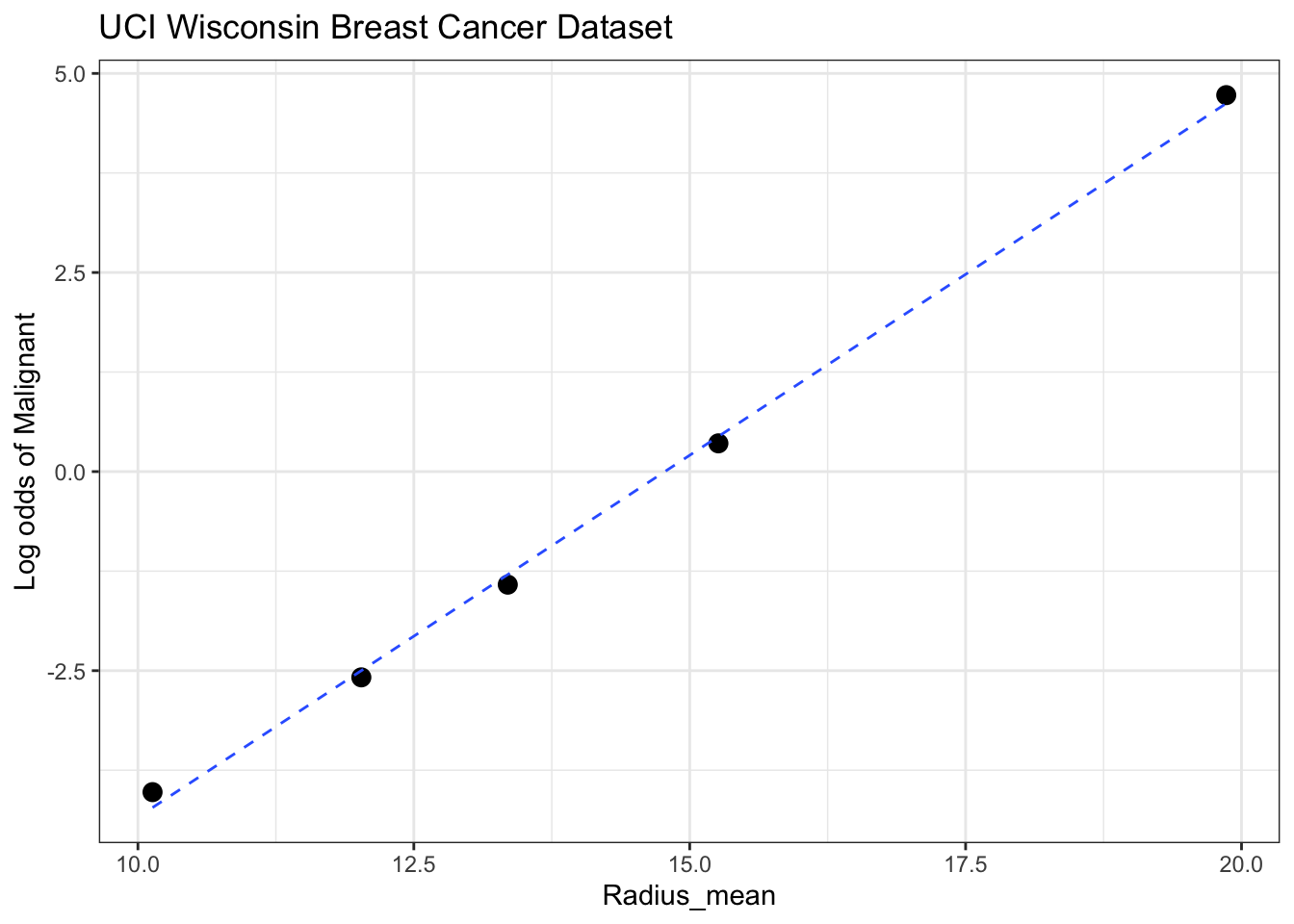
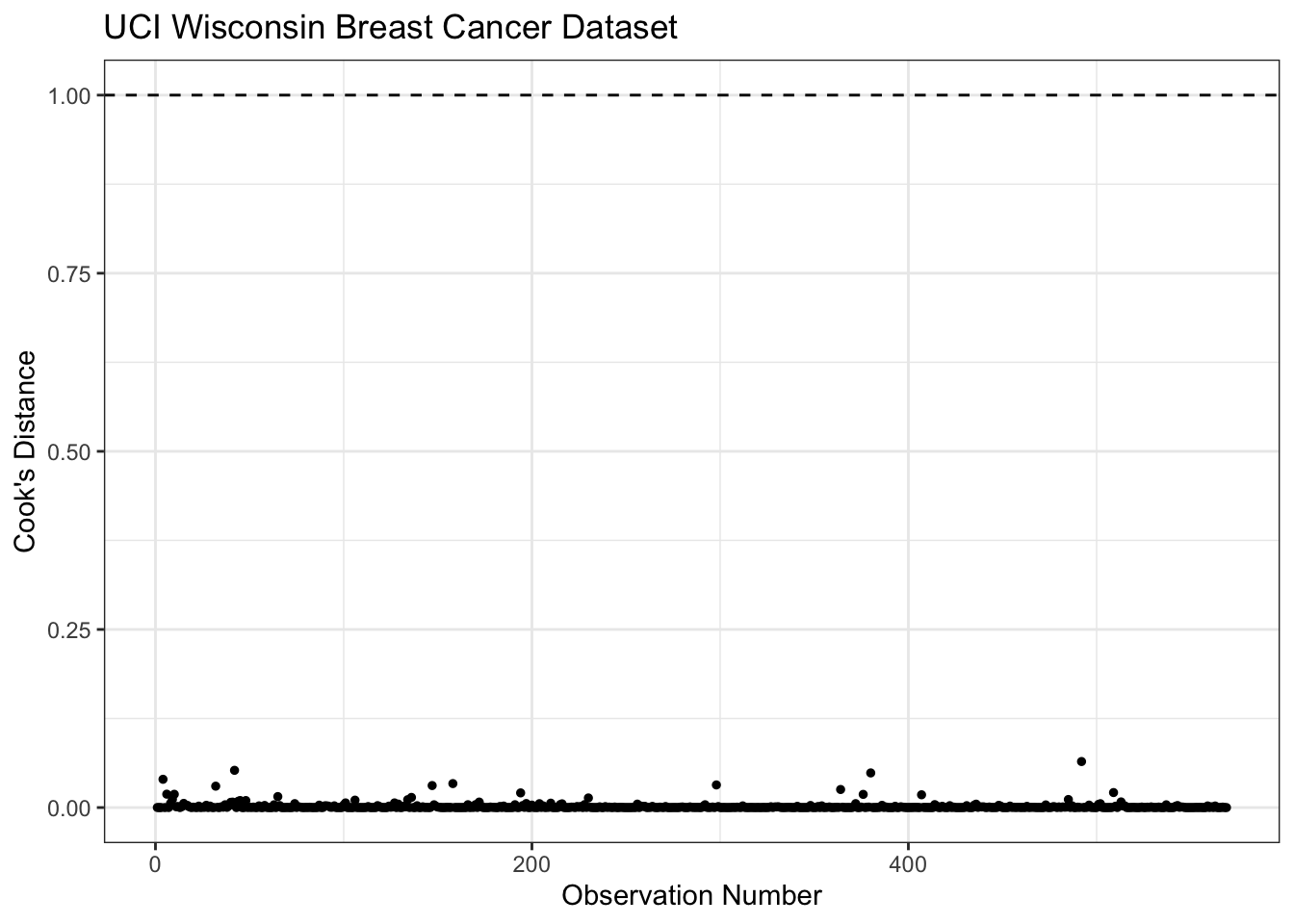
Rows: 569
Columns: 32
$ ID <int> 842302, 842517, 84300903, 84348301, 84358402, 8437…
$ Diagnosis <fct> M, M, M, M, M, M, M, M, M, M, M, M, M, M, M, M, M,…
$ Radius_mean <dbl> 17.990, 20.570, 19.690, 11.420, 20.290, 12.450, 18…
$ Texture_mean <dbl> 10.38, 17.77, 21.25, 20.38, 14.34, 15.70, 19.98, 2…
$ Perimeter_mean <dbl> 122.80, 132.90, 130.00, 77.58, 135.10, 82.57, 119.…
$ Area_mean <dbl> 1001.0, 1326.0, 1203.0, 386.1, 1297.0, 477.1, 1040…
$ Smoothness_mean <dbl> 0.11840, 0.08474, 0.10960, 0.14250, 0.10030, 0.127…
$ Compactness_mean <dbl> 0.27760, 0.07864, 0.15990, 0.28390, 0.13280, 0.170…
$ Concavity_mean <dbl> 0.30010, 0.08690, 0.19740, 0.24140, 0.19800, 0.157…
$ Nconcave_mean <dbl> 0.14710, 0.07017, 0.12790, 0.10520, 0.10430, 0.080…
$ Symmetry_mean <dbl> 0.2419, 0.1812, 0.2069, 0.2597, 0.1809, 0.2087, 0.…
$ Fractaldim_mean <dbl> 0.07871, 0.05667, 0.05999, 0.09744, 0.05883, 0.076…
$ Radius_se <dbl> 1.0950, 0.5435, 0.7456, 0.4956, 0.7572, 0.3345, 0.…
$ Texture_se <dbl> 0.9053, 0.7339, 0.7869, 1.1560, 0.7813, 0.8902, 0.…
$ Perimeter_se <dbl> 8.589, 3.398, 4.585, 3.445, 5.438, 2.217, 3.180, 3…
$ Area_se <dbl> 153.40, 74.08, 94.03, 27.23, 94.44, 27.19, 53.91, …
$ Smoothness_se <dbl> 0.006399, 0.005225, 0.006150, 0.009110, 0.011490, …
$ Compactness_se <dbl> 0.049040, 0.013080, 0.040060, 0.074580, 0.024610, …
$ Concavity_se <dbl> 0.05373, 0.01860, 0.03832, 0.05661, 0.05688, 0.036…
$ Nconcave_se <dbl> 0.015870, 0.013400, 0.020580, 0.018670, 0.018850, …
$ Symmetry_se <dbl> 0.03003, 0.01389, 0.02250, 0.05963, 0.01756, 0.021…
$ Fractaldim_se <dbl> 0.006193, 0.003532, 0.004571, 0.009208, 0.005115, …
$ Radius_extreme <dbl> 25.38, 24.99, 23.57, 14.91, 22.54, 15.47, 22.88, 1…
$ Texture_extreme <dbl> 17.33, 23.41, 25.53, 26.50, 16.67, 23.75, 27.66, 2…
$ Perimeter_extreme <dbl> 184.60, 158.80, 152.50, 98.87, 152.20, 103.40, 153…
$ Area_extreme <dbl> 2019.0, 1956.0, 1709.0, 567.7, 1575.0, 741.6, 1606…
$ Smoothness_extreme <dbl> 0.1622, 0.1238, 0.1444, 0.2098, 0.1374, 0.1791, 0.…
$ Compactness_extreme <dbl> 0.6656, 0.1866, 0.4245, 0.8663, 0.2050, 0.5249, 0.…
$ Concavity_extreme <dbl> 0.71190, 0.24160, 0.45040, 0.68690, 0.40000, 0.535…
$ Nconcave_extreme <dbl> 0.26540, 0.18600, 0.24300, 0.25750, 0.16250, 0.174…
$ Symmetry_extreme <dbl> 0.4601, 0.2750, 0.3613, 0.6638, 0.2364, 0.3985, 0.…
$ Fractaldim_extreme <dbl> 0.11890, 0.08902, 0.08758, 0.17300, 0.07678, 0.124…