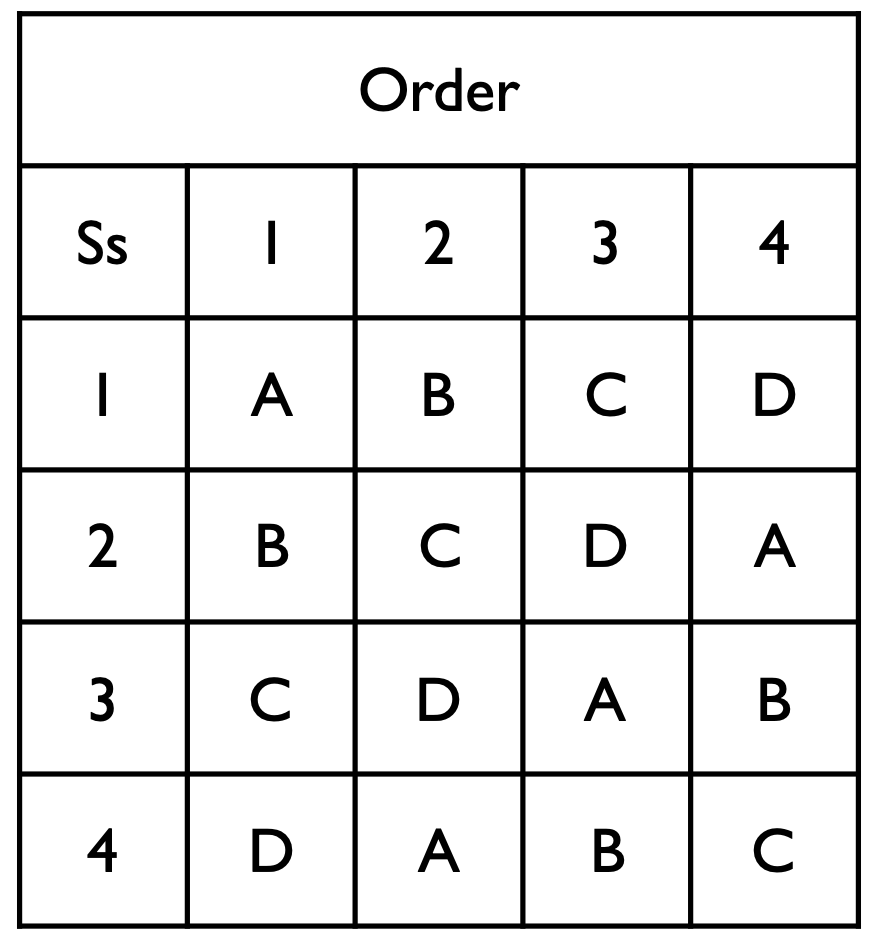
Code
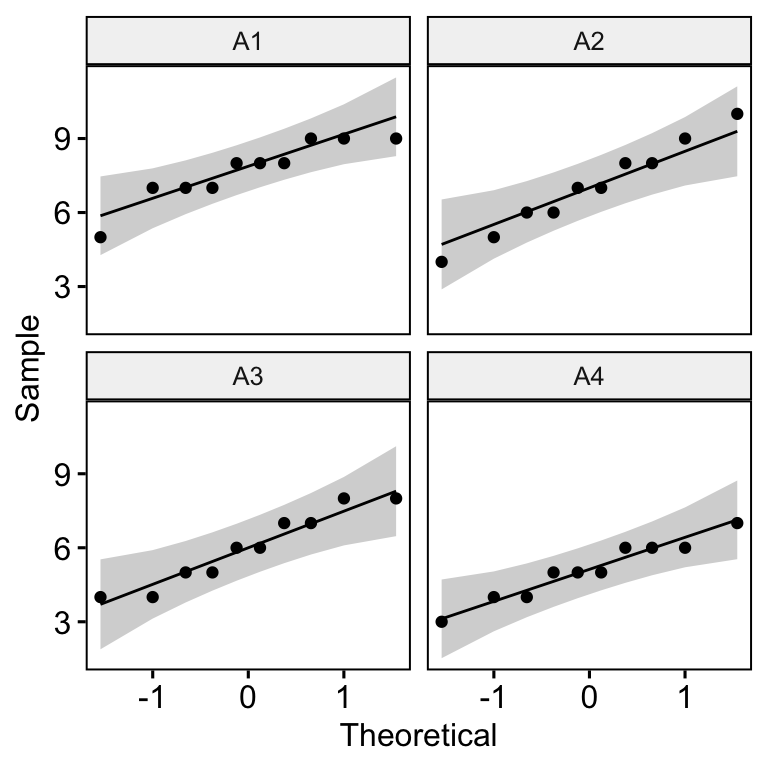
# A tibble: 10 × 5
Subject A1 A2 A3 A4
<fct> <dbl> <dbl> <dbl> <dbl>
1 1 8 10 7 5
2 2 9 9 8 6
3 3 7 5 8 4
4 4 9 6 5 7
5 5 8 7 7 6
6 6 5 4 4 3
7 7 7 6 5 4
8 8 8 8 6 6
9 9 9 8 6 5
10 10 7 7 4 5